How to run lcvplants in parallel
Source:vignettes/articles/Parallel-name-harmonization.Rmd
Parallel-name-harmonization.RmdAbout
Some users may want to use lcvplants functions to
perform taxonomic harmonization for many thousands of species and wish
to use the entire computational capacity available to speed up the
processing time. In this brief tutorial, we show how to run
lcvp_search or lcvp_fuzzy_search in parallel
in an efficient way to reduce computational time.
Should I run in parallel?
Before running the code in parallel, you should decide whether you need it. To decide that, you should take into account both the gain in processing time and the necessary time to adapt your code to run in parallel.
Check the following graph to decide whether do you think it is worth running the code in parallel. It shows the results of a test on a notebook using 3 cores and three different strategies (sequential, multicore, and multisession) to harmonize random samples of names in the TRY database of different sizes.

Required packages
Here, we are going to use the framework of the future
package to parallelize our code, as it can be easily adapted to users
running the code on different operational systems. So, make sure you
have the following packages installed and loaded.
The dataset
We are going to use here as an example a random sample of 100 names
from the Leipzig Catalogue of Vascular Plants database. In your code,
substitute the sps object for your vector of species
names.
Running in parallel
Number of cores
Before parallelizing your code, you need to decide what strategy you are going to use. Most users may want to parallelize the code on a local machine with several cores (via a multicore or multisession approach). For these users, the first thing is to decide how many cores on your local machine will be used for parallelization. You can check how many cores you have using the following code:
availableCores()
#> system
#> 4Decide how many of them you are going to use. Normally you may want to leave one out for other processes on your computer, but this is up to you.
cores <- availableCores() - 1Preparing the data
The second decision concerns how to divide the data to run the code
in parallel. Since all names could be searched independently, a user
could tell lcvp_search to harmonize each species name in a
different core. Although this may improve computational performance, it
is not the most efficient way to do that. The lcvplants
package was designed to optimize the computation over large vectors of
plant names and we should take advantage of that. So, the best way to
parallelize the code is to divide your data according to the number of
cores (or machines) available.
We run some tests on a workstation with 8 cores to harmonize random samples of names in the TRY database, see the results below:
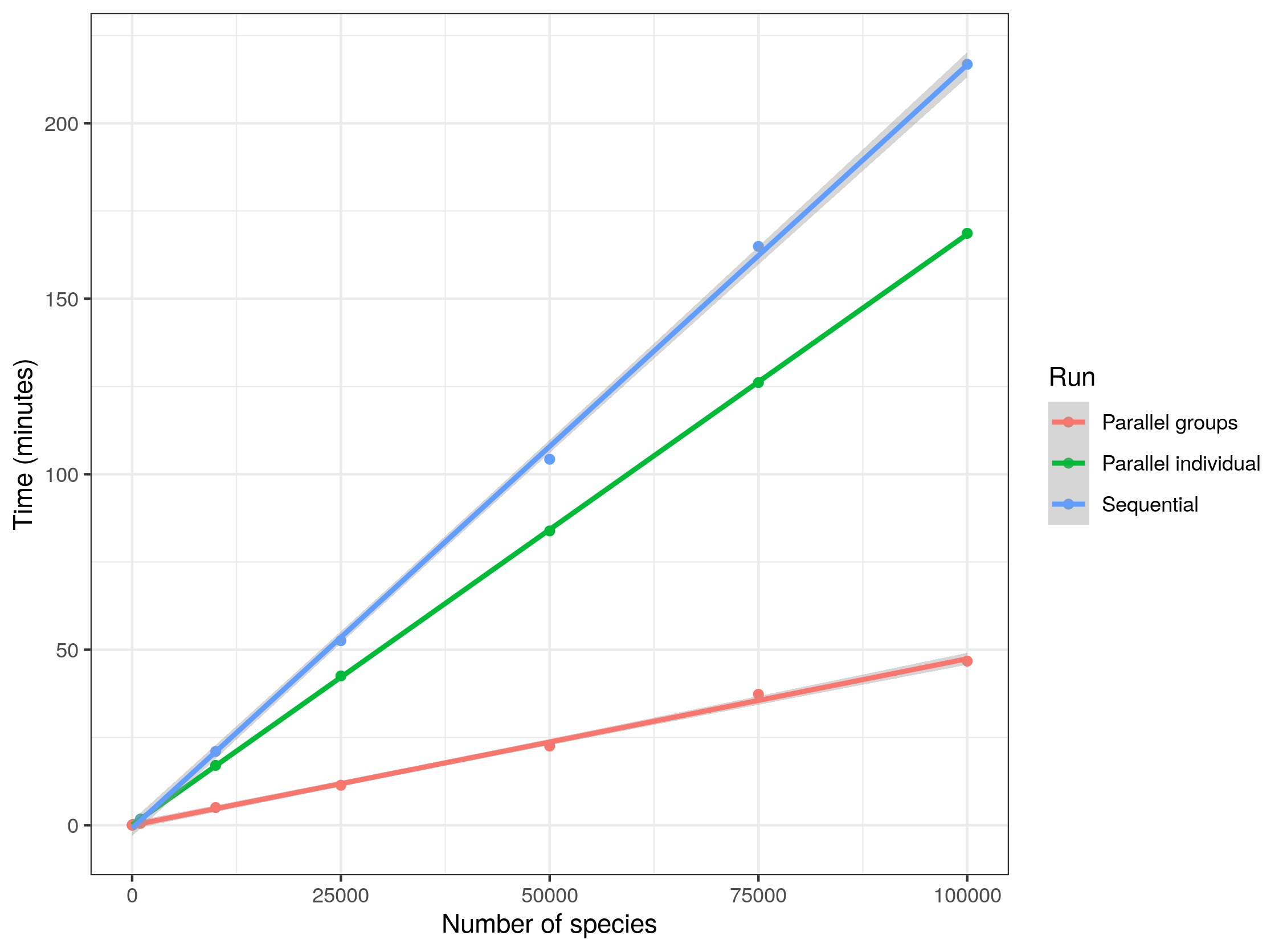
In other words, if you want to run your code using 3 cores, you
should divide the total dataset into 3 subsets, and then run
lcvp_search in 3 parallel cores. Use the following code to
divide your dataset.
Parallelization with the lapply approach
We can use the lapply approach applied by several
packages to parallelize our code here. In our example,
lapply can be used to apply the lcvp_search
function on our divided list of species names
(sps_list).
result <- lapply(sps_list, lcvp_search)The result will be also separated in a list with a length equal to
the original input list (in this case the object sps_list).
To combine the results, we can use the following code:
result_comb <- do.call(rbind, result)Notice now that the lapply runs a function over a list,
and each object of this list is processed independently. So, we could
tell R to process each element of the list in a different core. For
this, we will use the future_lapply function, but several
other functions based on the lapply to run in parallel are
available (e.g., parallel::mclapply or
parallel::parLapply). The structure to use
future_apply is the same as before.
plan(sequential)
result <- future_lapply(sps_list, lcvp_search)
result_comb <- do.call(rbind, result)Parallelization strategy
You may have noted that we used plan(sequential) before
the code. The function plan allows you to decide which
parallelization strategy to use. Several options are available:
sequential, multisession,
multicore, cluster, and remote
(see ?plan for details). If you are running the code on a
local machine, the faster option is to use the multicore
strategy (or forking approach). But this option does not work for
Windows machines (or in Rstudio), which in this case, the option
multisession (or socket approach) should be used.
You can check if the multicore option is available for you using the following code:
supportsMulticore()
#> [1] FALSEIf the answer is TRUE you can use the multicore option,
otherwise use the multisession.
strategy <- ifelse(supportsMulticore(),
"multicore",
"multisession")
plan(strategy, workers = cores)Then just run the code as before.
result <- future_lapply(sps_list, lcvp_search)
result_comb <- do.call(rbind, result)This same approach shown here can be used for
lcvp_fuzzy_search.